Leiden Early Drug Discovery & Development
Activity-based protein profiling for drug discovery
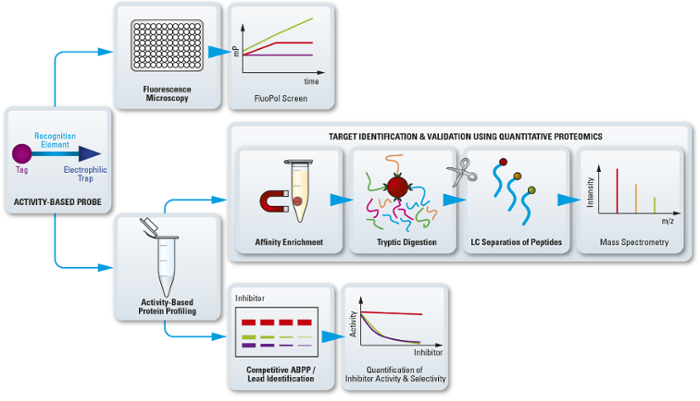
Activity-based protein profiling (ABPP, also termed chemical proteomics), is one of the pillars of chemical biology, and at LED3 we have taken it to the next level. ABPP allows the assessment of protein function in live cells and tissues, which means that the activity of a complete protein family can be measured under physiological conditions, such as in presence of protein-interaction partners, post-translational modifications, endogenous substrates and co-factors.
ABPP can accelerate drug discovery
In addition, ABPP can also show how protein activity is modulated by hits, leads and experimental drugs. This makes it a powerful tool in medicinal chemistry and a driving force in accelerating drug discovery.
We have used ABPP, for instance, to profile the off-target interaction landscape of the fatal experimental drug BIA 10-2474, which caused the death of a volunteer in a phase 1 clinical trial. We showed that this compound inhibited several lipases in the human brain and cortical neurons, which were not targeted by another safe fatty acid amide hydrolase inhibitor, suggesting that promiscuous lipase inhibitors may cause metabolic dysregulation. [1,2]
Taking ABPP further at LED3
ABPP also allows visualization of target engagement, thereby providing an essential correlate between target binding and a functional read-out in cellular and ultimately animal models. [3,4,5] We have been the first to successfully target a non-enzyme protein family by affinity-based protein profiling, i.e. G protein-coupled receptors. [6,7]
We were also the first to report a complete set of inhibitors and activity-based probes (ABPs) that report on proteasome activities in the context of human hematological cancers. [8] ABPP assays that make use of fluorescence polarization are amenable to a high-throughput format, and are employed to screen for new inhibitors, for instance for glycosidases involved in inherited disorders and cancer.[9,10]
Finding targets for antibiotics
ABPP not only allows us to identify target proteins of targeted covalent inhibitors, but also their exact interaction sites (residue-specific ABPP). We have tailored the underlying technologies for efficient identification of new druggable targets for antibiotics, and expanded the method to allow monitoring of aspartates, glutamates, histidines, tryptophanes, arginines and other amino acids. [11,12,13]
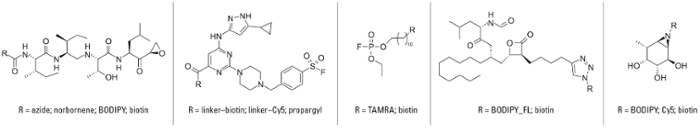
In addition, we provided a general roadmap for the unbiased evaluation of electrophile reactivity in the proteome. To date, we have assembled what is arguably the most comprehensive (and continuously expanding) activity-based probe collection worldwide, including ABPs selective for serine hydrolases, cysteine proteases, threonine proteases, glycosidases, ligases, lipases, amidases and kinases as well as for nine different amino acids.
References
- 1. A.C.M. van Esbroeck, A.P.A. Janssen, A.B. Cognetta III, D. Ogasawara, G. Shpak, M. van der Kroeg, V. Kantae, M.P. Baggelaar, F.M.S. de Vrij, H. Deng, M. Allarà, F. Fezza, Z. Lin, T. van der Wel, M. Soethoudt, E.D. Mock, H. den Dulk, I.L. Baak, B.I. Florea, G. Hendriks, L. De Petrocellis, H.S. Overkleeft, T. Hankemeier, C.I. De Zeeuw, V. Di Marzo, M. Maccarrone, B.F. Cravatt, S.A. Kushner and M. van der Stelt, Activity-based protein profiling reveals off-target proteins of the FAAH inhibitor BIA 10-2474, Science 2017, 356, 1084-1087. doi: 10.1126/science.aaf7497
- 2. E.J. van Rooden, B.I. Florea, H. Deng, M.P. Baggelaar, A.C.M. van Esbroeck, J. Zhou, H.S. Overkleeft and M. van der Stelt, Mapping in vivo target interaction profiles of covalent inhibitors using chemical proteomics with label-free quantification, Nat. Protoc. 2018, 13, 752-767. doi: 10.1038/nprot.2017.159
- 3. E.D. Mock, M. Mustafa, O. Gunduz-Cinar, R. Cinar, G.N. Petrie, V. Kantae, X. Di, D. Ogasawara, Z.V. Varga, J. Paloczi, C. Miliano, G. Donvito, A.C.M. van Esbroeck, A.M.F. van der Gracht, I. Kotsogianni, J.K. Park, A. Martella, T. van der Wel, M. Soethoudt, M. Jiang, T.J. Wendel, A.P.A. Janssen, A.T. Bakker, C.M. Donovan, L.I. Castillo, B.I. Florea, J. Wat, H. van den Hurk, M. Wittmer, U. Grether, A. Holmes, C.A.A. van Boeckel, T. Hankemeier, B.F. Cravatt, M.W. Buczynski, M.N. Hill, P. Pacher, A.J. Lichtman and M. van der Stelt, Discovery of a NAPE-PLD inhibitor that modulates emotional behavior in mice, Nat. Chem. Biol. 2020, 16, 667-675. doi: 10.1038/s41589-020-0528-7
- 4. D. Ogasawara, H. Deng, A. Viader, M.P. Baggelaar, A. Breman, H. den Dulk, A.M.C.H. van den Nieuwendijk, M. Soethoudt, T. van der Wel, J. Zhou, H.S. Overkleeft, M. Sanchez-Alavez, S. Mo, W. Nguyen, B. Conti, X. Liu, W.-S. Liu, B.F. Cravatt and M. van der Stelt, Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition, Proc. Natl. Acad. Sci. USA 2016, 113, 26-33. doi: 10.1073/pnas.1522364112
- 5. E. van Meel, E. Bos, M.J.C. van den Lienden, H.S. Overkleeft, S.I. van Kasteren, A.J. Koster and J.M.F.G. Aerts, Localization of active endogenous and exogenous beta-glucocerebrosidase by correlative light-electron microscopy in human fibroblasts, Traffic 2019, 20, 346-356. doi: 10.1111/tra.12641
- 6. M. Soethoudt, S.C. Stolze, M.W. Westphal, L. van Stralen, A. Martella, E.J. van Rooden, W. Guba, Z.V. Varga, H. Deng, S.I. van Kasteren, U. Grether, A.P. IJzerman, P. Pacher, E.M. Carreira, H.S. Overkleeft, A. Ioan-Facsinay, L.H. Heitman and M. van der Stelt, Selective photoaffinity probe that enables assessment of cannabinoid CB2 receptor expression and ligand engagement in human cells, J. Am. Chem. Soc. 2018, 140, 6067-6075. doi: 10.1021/jacs.7b11281
- 7. X. Yang, T.J.M. Michiels, C. de Jong, M. Soethoudt, N. Dekker, E. Gordon, M. van der Stelt, L.H. Heitman, D. van der Es and A.P. IJzerman. An Affinity-Based Probe for the Human Adenosine A2A Receptor. J Med Chem. 2018, 61, 7892-7901. doi: 10.1021/acs.jmedchem.8b00860
- 8. G. de Bruin, B. T. Xin, M. Kraus, M. van der Stelt, G.A. van der Marel, A.F. Kisselev, C. Driessen, B.I. Florea and H.S. Overkleeft, A set of chemical tools to visualize and control human (immuno)proteasome activities, Angew. Chem. Int. Ed. 2016, 55, 4199-4203. doi: 10.1002/anie.201509092
- 9. D. Lahav, B. Liu, R.J.B.H.N. van den Berg, A.M.C.H. van den Nieuwendijk, T. Wennekes, A. T. Ghisaidoobe, I. Breen, M.J. Ferraz, C.-L. Kuo, L. Wu, P.P. Geurink, H. Ovaa, G A. van der Marel, M. van der Stelt, R. G. Boot, G. J. Davies, J.M.F.G. Aerts and H.S. Overkleeft, A fluorescence polarization activity-based protein profiling assay in the discovery of potent, selective inhibitors for human non-lysosomal glucosylceramidase, J. Am. Chem. Soc. 2017, 139, 14192-14197. doi: 10.1021/jacs.7b07352
- 10. Z. Armstrong, C.-L. Kuo, D. Lahav, B. Liu, R. Johnson, T.J.M. Beenakker, C. de Boer, C.-S. Wong, E.R. van Rijssel, M.F. Debets, B.I. Florea, C. Hissink, R.G. Boot, P.P. Geurink, H. Ovaa, M. van der Stelt, G. A. van der Marel, J.D.C. Codée, J.M.F.G. Aerts, L. Wu, H.S. Overkleeft and G.J. Davies, Manno-epi-cyclophellitols enable activity-based protein profiling of human -mannosidases and discovery of new Golgi mannosidase II inhibitors, J. Am. Chem. Soc. 2020, 142, 13021-13029. doi: 10.1021/jacs.0c03880
- 11. P.R.A. Zanon, L. Lewald, S.M. Hacker, Isotopically Labeled Desthiobiotin Azide (isoDTB) Tags Enable Global Profiling of the Bacterial Cysteinome, Angew. Chem. Int. Ed. 2020, 59, 2829-2836. doi: 10.1002/anie.201912075
- 12. K. Bach, B. Beerkens, P.R.A. Zanon, S.M. Hacker, Light-activatable, 2,5-Disubstituted Tetrazoles for the Proteome-wide Profiling of Aspartates and Glutamates in Living Bacteria, ACS Cent. Sci. 2020, 6, 546-554. doi: 10.1021/acscentsci.9b01268
- 13. P.R.A. Zanon, F. Yu, P. Musacchio, L. Lewald, M. Zollo, K. Krauskopf, D. Mrdović, P. Raunft, T.E. Maher, M. Cigler, C. Chang, K. Lang, F.D. Toste, A.I. Nesvizhskii and S.M. Hacker, Profiling the Proteome-Wide Selectivity of Diverse Electrophiles, ChemRxiv 2021, doi: 10.26434/chemrxiv.1418656

