Macromolecular Biochemistry
Research
Research at the Macromolecular Biochemistry group is comprised of the following research themes:
Enzyme evolution, structure, dynamics and activity (Marcellus Ubbink)
Enzymes are beautiful catalysts responsible for accelerating and regulating nearly all biochemical reactions. We want to understand how the protein matrix can achieve its function. We aim to decipher the interplay of residues in and around the active site with the substrate, as well as the role of the residues farther away in maintaining the correct structure. In particular, we are interested to understand how structure and dynamics of the protein influence the catalytic process and how it evolved over billions of years.
Chromatin organisation & dynamics (Remus Dame)
The genomic DNA of every organism is organized and compacted in order to fit inside the cell. This is achieved by the joint action of chromatin proteins that fold the genome. Our focus is on the chromatin proteins from bacteria and archaea, with a special focus on how ancestral chromatin proteins from archaea have evolved into the chromatin proteins that shape today’s eukaryotic genomes. Genome folding is tightly interconnected with transcription, with genes in certain regions being silenced, while others are highly transcribed. Our interest lies in understanding how chromatin proteins act on DNA and how they regulate transcription. We investigate the activity of these proteins in vitro as well as in vivo using biochemical and state-of-the-art biophysical approaches. Read more
Membrane proteins biohybrids (Lars Jeuken)
We aim to develop novel biophysical approaches to study membrane proteins in bacterial bioenergetics and exploit them in biotechnology. We study the redox activity of respiratory-chain enzymes in a native-like model membrane environments to elucidate their interaction with, for instance, natural lipophilic quinones or novel antibiotics. Read more We combine our membrane proteins with with non-biological materials, creating ‘hybrid’ materials with potential applications in synthetic cells (Read more), semi-artificial photosynthesis (Read more) and biotechnology.
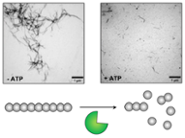
Molecular chaperones in neurodegeneration (Anne Wentink)
We investigate the role of molecular chaperones in the formation, clearance and propagation of toxic protein aggregates, so-called amyloid fibres, commonly found in the brains of Alzheimer’s and Parkinson’s disease patients.
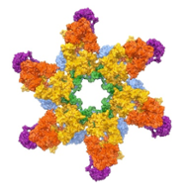
Secretion in pathogenic mycobacteria (Sebastian Geibel)
We investigate how pathogenic bacteria take up nutrients and export virulence factors across their cell envelope. The lab combines structural (cryo-EM, X-ray crystallography), biochemical and microbiological methods to gain insights into the structure and function of the bacterial systems that mediate these processes.
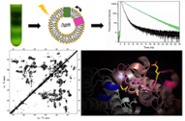
Structure and regulation in photosynthesis (Anjali Pandit)
We combine advanced spectroscopy and protein biochemistry to reveal molecular mechanisms of photosynthesis and we are interested in design of artificial proteins that mimic photosynthetic tasks. Ultimately, this information may be used to develop species that convert more light to biomass or for new concepts in artificial photosynthesis.
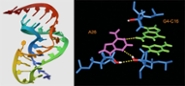
RNA structure and function (Rene Olsthoorn)
We study the role of RNA structures in viral and bacterial infections and genetic disorders with a strong focus on RNA pseudoknots and G-quadruplexes. Activities include RNA structure prediction and characterization by biochemical methods as well as functional analyses using in vitro and in vivo model systems. Additionally, methods are developed to screen small molecule drugs that target pathogenic RNAs.
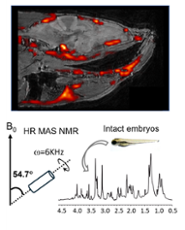
Neuroimaging and Metabolic Mechanisms (Alia)
We investigate the mechanisms underlying a spectrum of brain diseases using cutting-edge MRI and NMR techniques, coupled with zebrafish as a versatile model system. We also apply and tailor HR-MAS NMR approaches to investigate how neurotoxins and environmental pollutants, such as nanoplastics and PFAS, affect metabolic pathways and drive toxicity.
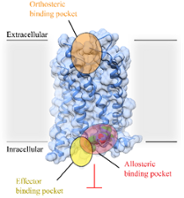
Structural Biology of GPCRs (Steffen Brünle)
We investigate the molecular mechanisms of cell signalling proteins using a range of methodologies in Structural Biology (single-particle cryo-EM and X-ray crystallography), Biochemistry, and Biophysics. We particularly focus on G-protein coupled receptors (GPCRs) and explores novel concepts in GPCR drug discovery. Read more
banner image: a) Evolution: β-lactamase; b) DNA packaging: archaeal histone; c) Bioenergetics: terminal oxidase; d) RNA structure: retrovirus pseudoknot; e) Photosynthesis: LHCII