Research project
Understanding plant transgenesis
How is Pol θ responsible for T-DNA integration, and how do other DNA double-strand break repair pathways interact with Pol θ? How may we manipulate T-DNA integration to stimulate error-free integration at a predetermined genomic site?
- Duration
- 2017 - 2020
- Contact
- Marcel Tijsterman
- Funding
-
 NWO
NWO
- Partners
Leiden University Medical Center (LUMC), Genome stability group

Unravelling the molecular mechanisms that underlie plant genetic modification.
In nature, Agrobacterium tumefaciens genetically modifies plants by inserting T-DNA, containing tumor-inducing genes, into their genomes, causing crown gall disease. In the laboratory, tumor-inducing genes can be replaced by any desired sequence, making genetic transformation by Agrobacterium the most popular tool to create genetically modified plants.
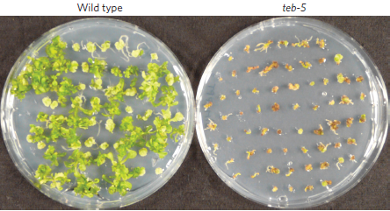
In 2016, we discovered, in collaboration with the Genome Stability Group at the LUMC, that T-DNA integration requires Polymerase Theta (Pol θ). DNA double-strand break repair by Pol θ captures T-DNA into genomic breaks. Random integration events, which constitute almost all events, are created by Pol θ. Random integration of T-DNA heavily impacts the function of integrated T-DNA, and impacts the genomic environment. Therefore, it represents a safety risk in genetically modified plants.
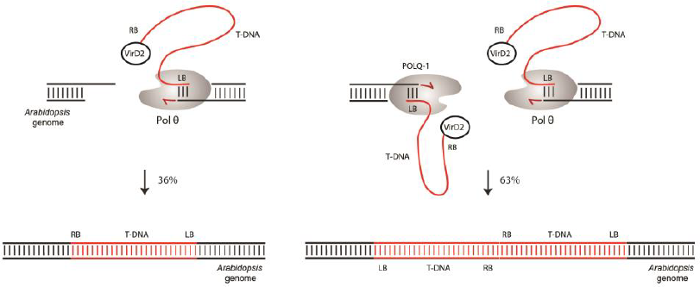
Current work aims to unravel the context in which Pol θ operates in T-DNA integration. We study interactions between Pol θ and other double-strand break repair pathways and their effects on the frequency of T-DNA integration, the molecular outcome of integration events, and explore possibilities to steer the T-DNA integration process towards error-free gene targeting.
- Van Kregten et al., T-DNA integration in plants results from polymerase-θ-mediated DNA repair, Nat. Plants, 2016
