
Cleaning up tuberculosis and salmonella infections
The cellular recycling system in zebrafish is capable of eating harmful bacteria and thus resist infections such as tuberculosis and salmonellosis. That is written by Leiden biologists from the group of Annemarie Meijer. Stimulating this form of defence could be used in new treatment methods against infectious diseases. The researchers publish their results in two leading journals, Autophagy and PLOS Pathogens.
Eating bacteria
Some of the world’s most dangerous infectious diseases are caused by bacteria that can replicate inside cells of the immune system, especially in so-called macrophages. It is already known that macrophages can defend themselves by eating these pathogens. This reaction is known as autophagy, which literally means ‘eating oneself’. In healthy organisms, this functions as a recycling system to clean up unwanted and damaged components of cells. However, during an immune response, this system is used to capture and break down microbial invaders. Stimulating this autophagy defence could, therefore, be a possible new treatment strategy against infectious diseases. The new research by Meijer and her colleagues in zebrafish models for tuberculosis and salmonellosis supports this idea.
Tuberculosis
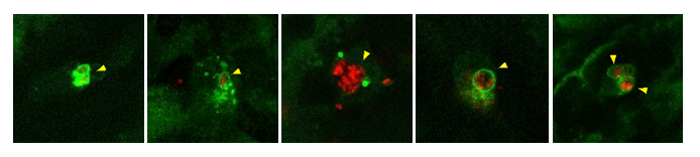
The research in PLOS Pathogens is about intruders from the family of mycobacteria. Different species of mycobacteria cause tuberculosis in humans and fish. They can replicate inside vesicles of infected macrophages, but they can also escape to another part of the cell, the cytoplasm. Here, they become targets of the autophagy defense. The Leiden biologists illustrated this using advanced microscopy and fluorescent autophagy proteins in zebrafish larvae. This showed that two autophagy receptors in zebrafish, p62 and Optineurin, are both required for defense against escaping bacteria.
This works as follows: escaped bacteria are marked in the cytoplasm with a compound called ubiquitin. The receptors recognize this signal and pick up a bacterium. They then deliver it to the autophagy machinery. ‘It turned out that fish without the receptors are much more susceptible to infection’, explains Meijer. ‘Fish that make more p62 or Optineurin turned out to be more resistant to infection.’
Salmonellosis
In the journal Autophagy the researchers focus on salmonella. Different variants of Salmonella enterica cause gastrointestinal infections (salmonellosis) or more serious systemic infections, like typhoid fever. With these pathogens, the researchers discovered an extra mechanism. They found that vesicles with salmonella are marked by a protein from the autophagy machinery, called Lc3. This happened shortly after the bacteria are engulfed by macrophages. ‘It is as if a ring of Lc3 forms around the bacteria’, says Meijer. ‘We think this helps to prevent the escape of the bacteria and stimulates fusion with digestive vesicles.’ The marking of infected vesicles is called LAP: Lc3-associated phagocytosis. The results identify LAP as a major host protective pathway in macrophage defence against Salmonella during systemic infection.
Anti-infection
The results of both studies encourage further research into autophagy defence. Meijer: ‘By stimulating this form of defence, we could possibly treat infections that are resistant to antibiotics. This can be done with the help of drugs that target proteins that can stimulate the autophagy process.’
Sources:
