Research project
Unspinning chromatin: molecular mechanisms of chromatin remodeling
How do you fit two meters of DNA in a tiny compartment and at the same time are able to access the right parts of it at the right times?
- Funding
- NWO Vidi
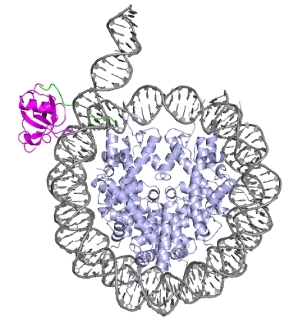
Chromatin is the complex of DNA and histone proteins that is needed to pack the eukaryotic genome into the cell nucleus. It ensures the extremely efficient storage of DNA, but at the same time it creates a barrier for access to the DNA in all essential nuclear processes, such as DNA transcription, replication, and repair. Chromatin remodeling is key in lifting, and reinstalling this barrier. Remodeling is therefore at the heart of chromatin biology and its defects are closely linked to developmental diseases and oncogenesis.
Because of its highly dynamic nature and the size of the molecules involved, the molecular mechanisms of chromatin remodeling are challenging to study experimentally. Dr. van Ingen proposes to exploit the exquisite sensitivity of nuclear magnetic resonance (NMR) towards molecular structure and dynamics, to study the remodeling of the fundamental repeating units of chromatin, nucleosomes, in atomic detail.
Recently, I have established that state-of-the art NMR methods can be applied to elucidate the structure of nucleosome-protein complexes. I will now capitalize on this to develop new methodology, combining solution and solid-state NMR techniques, to study the structural and dynamical basis of remodeling. I will focus on remodeling by incorporation of the highly divergent histone H2A.Bbd and by an energy-driven molecular machine that actively repositions nucleosomes, the imitation-switch remodeler Isw1.
These studies will offer unique mechanistic insights in chromatin remodeling, and yield novel NMR procedures that open major possibilities for the high-resolution analysis of other chromatin-associated systems, which will be valuable in the design of therapeutics and our fundamental understanding of chromatin in health and disease.
