Research project
Staying Ahead of the Virus
In STAYAHEAD data-intensive approaches are being developed to ”decode the human immunome” with a focus on a global vaccine strategy. They have developed a rapid mass spectrometric test to analyse in real-time large numbers of variants of SARS-CoV-2 and the host immune response, and use these data to predict potential high-risk variants.
- Duration
- 2022 - 2025
- Contact
- Thomas Hankemeier
- Partners
All partners involved in STAYAHEAD can be found on the project site: https://www.health-holland.com/project/2023/2022/staying-ahead-virus
Rapid SARS-CoV-2 detection and variant characterization using mass spectrometry coupled to AI predictions of variants of high risk: Staying ahead of the virus (STAYAHEAD)
The COVID-19 pandemic focused attention on the mechanisms of emergence and spread of SARS-CoV-2 variants that have increased transmissibility, virulence, or potential to escape the immune response induced by earlier infection or vaccination.
In this 3 year project (funded by Health-Holland and initiated October 2022 and building on a mass spectrometric HT assay for viral diagnostics developed during the pandemic) The aim is to develop:
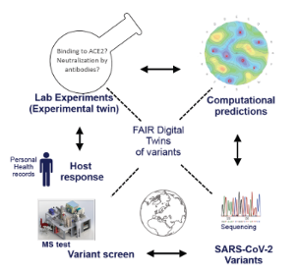
(1) a modular Mass-Spectrometry (MS)-based platform to analyze peptide fragments of the SARS-CoV-2 NCAP and spike proteins for the diagnosis and characterization of SARS-CoV-2 infections via variant detection as well as the characterization of patient immune response to SARS-CoV-2 infection via tracking of the clinical course of disease;
(2) complementary large-scale computational predictions of protein variant structures covering all know variants but also extending into the much larger space of possible sequences (scaling to several billions of virtual sequences). These large and complex data will be rendered as FAIR Knowledge Graphs that can be used by Artificial Intelligence systems to predict protein variants that are likely to emerge in the near future and their associated health risks including their potential to escape the immune response acquired by vaccination or earlier infections;
(3) As proof-of-concept, they will use in vitro models to test if predicted high risk variants can indeed escape antibodies due to earlier infections or vaccination. Core to this project is the development of an open data knowledge graph based on FAIR Digital Twins (FDT) of individual protein variants.
The FDT allows automated management of the (i) real-world, (ii) experimental and (iii) computational data generated in this project and from external sources. The MS testing platform, real-world data collection, and prediction/validation platforms are synergistically linked via an AI feedback loop driven by the FAIR Knowledge Graphs providing decision-intelligence for accelerating the research cycle to meet the urgency of pandemic outbreaks. Ultimately, any novel variant that is detected or predicted to be of concern will automatically trigger software updates to the network of globally distributed MS platforms, allowing real-time, worldwide detection of novel variants of the ever-evolving SARS-CoV-2 virus. As this strategy can be applied to any viral infection, this kick-start project will be key to pandemic preparedness roadmaps.
This project represents the first step to building a FAIR Knowledge Graph-based data collection infrastructure in the Leiden Academic Center for Drug Research data collection, and an early contribution to the international Human Immunome Project.
https://www.health-holland.com/project/2023/2022/staying-ahead-virus
