Research project
Chromatin organisation & dynamics
The genomic DNA of every organism is organized and compacted in order to fit inside the cell. This is achieved by the joint action of numerous architectural proteins that aid in folding the genome. Genome folding is tightly interconnected with transcription, with genes in certain regions being silenced, while others are highly transcribed. Our interest lies in understanding how architectural proteins act on DNA and how they regulate transcription. We investigate the activity of these proteins in vitro as well as in vivo using biochemical and state-of-the-art biophysical approaches.
- Contact
- Remus Dame
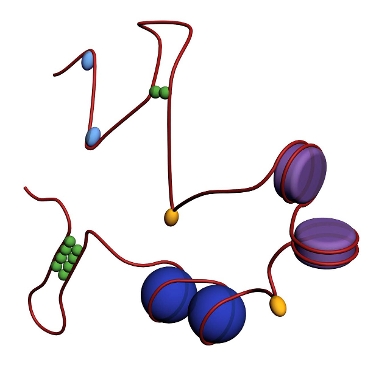 The genetic information stored in the DNA of organisms from all three domains of life is folded into a structure that is compact, yet functionally organised in a way that permits genome activity. In bacteria and archaea the genome is not membrane enclosed and “free floating” yet centrally positioned in the cytoplasm, whereas in eukaryotes the genome is located in a dedicated organelle, the nucleus and surrounded by the nuclear membrane. Independent of these cytological differences, all genomes are shaped by the action of abundant proteins with architectural function. This folded structure including the associated architectural proteins is referred to as chromatin. Architectural proteins, although minimally conserved in terms of structure and sequence among kingdoms, act upon DNA using conserved mechanisms (Luijsterburg et al., Crit Rev Biochem Mol Biol, 2008). Well-known examples are the eukaryotic and archaeal histones that wrap DNA and the nucleoid-associated proteins in bacteria and archaea that bend and bridge DNA.?
The genetic information stored in the DNA of organisms from all three domains of life is folded into a structure that is compact, yet functionally organised in a way that permits genome activity. In bacteria and archaea the genome is not membrane enclosed and “free floating” yet centrally positioned in the cytoplasm, whereas in eukaryotes the genome is located in a dedicated organelle, the nucleus and surrounded by the nuclear membrane. Independent of these cytological differences, all genomes are shaped by the action of abundant proteins with architectural function. This folded structure including the associated architectural proteins is referred to as chromatin. Architectural proteins, although minimally conserved in terms of structure and sequence among kingdoms, act upon DNA using conserved mechanisms (Luijsterburg et al., Crit Rev Biochem Mol Biol, 2008). Well-known examples are the eukaryotic and archaeal histones that wrap DNA and the nucleoid-associated proteins in bacteria and archaea that bend and bridge DNA.?
Chromatin is a dynamic entity. Its basic shape is imposed by the proteins mentioned above, but it is subject to continuous remodeling to live up to the demands of genes being expressed or repressed. It is remodeled actively in eukaryotes by motor enzymes that can move or displace nucleosomes, in addition to remodeling mechanisms more ubiquitous in bacteria and archaea that are based on the interplay between multiple architectural proteins. Remodeling of chromatin is also mediated by chemical modifications of the DNA, the architectural proteins that shape it or physicochemical cues.
An emerging theme in the field is how genome organisation in unicellular organisms is affected by cellular environment and growth conditions. Models suggest that in bacteria and archaea there are direct effects of physicochemical factors such as osmolarity, temperature and pH on the action of chromatin proteins, in addition to indirect effects by modulating the expression ratios of different types of chromatin proteins. In this light, my group has initiated studies on the effects of osmolarity as well as temperature on protein-DNA interactions in single-molecule and ensemble in vitro studies (Vreede & Dame, Biophys J., 2012; Driessen et al. Submitted.; van der Valk et al., In preparation.). Also, my group is investigating the (architectural) interplay between different types of chromatin proteins in vitro (Laurens et al., Nature Comm, 2012; van der Valk et al., In preparation.). In addition to dissecting these mechanistic aspects of gene regulation, we are interested in the roles and physiological consequences of the products of genes regulated by chromatin proteins. In that light, we have been studying the E. coli CRISPR/Cas system (Westra et al., Mol Cell, 2012; Westra et al., RNA biol, 2012) and cell motility (Shahapure et al., Mol Mic, 2014).
Our current aim is to obtain an integrated picture of the interplay between spatial genome organization and DNA transactions in general and transcription in specific. The studies of molecular aspects of genome organisation determined in vitro have yielded models for their in vivo functionality, which urgently require testing in the cellular context. Of particular interest are the models for global genome organization based on DNA looping in which a central role is played by the H-NS (E. coli) and Alba (Sulfolobus sp.) proteins. Based on the known role of H-NS as transcriptional repressor (and the proposed analogous role of Alba) it is expected that the bridged structure at the base of genomic loops is the region responsible for repression. Thus genome organisation and transcription regulation are likely intimately coupled. As transcription of many genes and operons responds to environmental changes and as these are often mediated by chromatin proteins, it is expected that 1) the architectural interplay of such proteins and/or 2) their direct response to physicochemical changes determines loop formation and dissolution leading to altered transcription levels. We are investigating these two possibilities using our longstanding experience with advanced in vitro techniques. More importantly, we are using and developing new approaches to investigate genome organisation and its coupling to transcription in vivo. Specifically, we are employing these approaches to investigate the remodeling of genomic regions (i.e. genes or operons up/down regulated following a stimulus) in 3 dimensions to quantitatively dissect the coupling between stimulus, local genome conformation and gene activity. The genomic regions of our interest contain H-NS regulated operons known to respond to the physicochemical cues described above. Such cues are for instance encountered when a bacterium is colonizing a host organism. As such they are key determinants of bacterial pathogenicity and very important in relation to human health.
Key publications
- Driessen, R.P.C., H. Meng, G. Suresh, R. Shahapure, G. Lanzani, U.D. Priyakumar, M.F. White, H. Schiessel, et al., "Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends", Nucleic Acids Research, vol. 41, no. 1, pp. 196-205, Jan, 2013. DOI: 10.1093/nar/gks1053
- Laurens, N., R.P.C. Driessen, I. Heller, D. Vorselen, M.C. Noom, F.J.H. Hol, M.F. White, R.T. Dame, et al., "Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA", Nature Communications, vol. 3, Dec, 2012. DOI: 10.1038/ncomms2330
- Noom, M.C., W.W. Navarre, T. Oshima, G.J.L. Wuite, R.T. Dame, "H-NS promotes looped domain formation in the bacterial chromosome", Current Biology, vol. 17, no. 21, pp. R913-R914, Nov 6, 2007. DOI: 10.1016/j.cub.2007.09.005
- Dame, R.T., M.C. Noom, G.J.L. Wuite, "Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation", Nature, vol. 444, no. 7117, pp. 387-390, Nov 16, 2006. DOI: 10.1038/nature05283
- van Noort, J., S. Verbrugge, N. Goosen, C. Dekker, R.T. Dame, "Dual architectural roles of HU: Formation of flexible hinges and rigid filaments", Proceedings of the National Academy of Sciences, vol. 101, issue 18, pp. 6969 - 6974, 05/2004. DOI: 10.1073/pnas.0308230101
