
How mammoth poop contributes to antibiotics research
PhD student Doris van Bergeijk brought 40,000-year-old bacteria from mammoth poop back to life. She hopes to find new information that can help research at the Institute of Biology Leiden into antibiotics and antibiotics resistance. Read about it on European Antibiotic Awareness Day, 18 November.
‘I find it very extraordinary that we can work with a mammoth stool sample in our lab. It is almost 40,000 years old,' says PhD student Doris van Bergeijk. Van Bergeijk conducts research into antibiotic-producing bacteria at the research group of Professor Gilles van Wezel at the Institute of Biology Leiden. At first glance it might not be obvious to work with mammoth poop, but Van Wezel and his team knew how to put it to good use.

Hibernation
The sample emanated from a very well preserved mammoth found in the permanently frozen ground – permafrost – of the island of Malo Lyakhovsky in the Arctic Ocean near Russia. In 2015, physician-microbiologists Dries Budding and Bas Wintermans took samples of animal tissues and intestinal contents back to the Netherlands. The mammoth turned out to be 40,000 years old, and so were the traces of bacteria that were found in the stool. Some types of bacteria, like many fungi and some plants, create spores to survive. The spores contain the genetic material and are resistant to unfavourable conditions such as heat and dehydration. Under the right conditions they can develop into cells that can grow and divide. Together with postdoctoral researcher Mia Urem Van Bergeijk succeeded in germinating 40,000-year-old spores: ‘We sort of woke them up from a very long hibernation.’
The same but different
Van Bergeijk is particularly interested in Actinobacteria: ‘Actinobacteria are able to produce lots of different antibiotics. We put the spores on specific nutrient media to see where they would grow best.’ It worked; a variety of Actinobacteria were retrieved. The bacteria were then examined by sequencing them – a method to read the DNA code. This was done in collaboration with professor Herman Spaink and assistant professor Victor Carrión from IBL, and the company Future Genomics Technologies in Leiden. That yielded interesting information: ‘The DNA of the bacterial strains from the mammoth sample seems to differ surprisingly from the DNA of known strains, although they are not completely new bacteria,’ explains Van Bergeijk. ‘We don't know if those differences are because they are 40,000 years old or because they come from permafrost. We are now investigating that.’
Turn it on
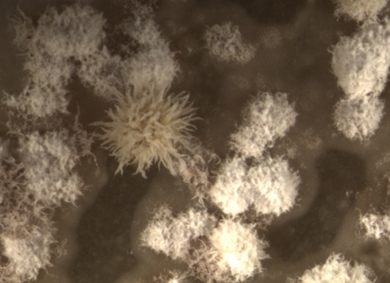
One bacterial strain that Van Bergeijk has cultivated has her particular interest: ‘The DNA research shows that this so-called streptomycete has the ability to produce potentially very interesting molecules. The bacterium has a coral-like structure, which looks amazing.’ According to Van Bergeijk it is difficult to ‘turn on’ the antibiotics-production of these bacteria in a laboratory setting. When and under what circumstances bacteria will produce antibiotics is one of the themes within the Van Wezel research group, where Van Bergeijk works. ‘The bacteria that we have cultivated from the mammoth stool may potentially offer clues for finding new antibiotics and for understanding the evolution of these species; can those bacteria make something the same species can no longer do, or vice versa?’ With a little over a year left until the end of her PhD, Van Bergeijk still has enough questions to which she hopes to find answers.
Header image: Streptomycete producing drops of antibiotics. (Photo: Doris van Bergeijk)
