Background
AMR, or AntiMicrobial Resistance, starts to develop immediately after introduction of a new antimicrobial agent. Therefore, a continuous introduction of novel antibiotics is required.
Brief history
After the discovery of penicillin by Alexander Fleming in 1928, it took until the 1940s-1950s before antibiotics became commercially available for the general population. From that moment on, many more antibiotics were discovered, and they were extensively used to combat infections. It was even believed that mankind had conquered infectious diseases. However, immediately after introduction of new antibiotics, resistance started to develop, a phenomenon already observed by Fleming. The widespread use of antibiotics has led to the development of many bacterial species and strains that are resistant to one or even multiple antibiotics.
The development of antimicrobial resistance (AMR) motivated the search for novel antibiotics. Until the 1980s many new compounds were introduced. However, in the classical screens replication started to occur: the screens did not yield novel antibiotics, but only molecules that are identical or similar to known antibiotics. The pharmaceutical industry then tried different approaches, such as combinatorial chemistry, genomics-based target identification and impressive screenings of compound libraries. The limited success of these efforts, and the large costs of research, caused pharmaceutical companies to cease their activities in the field of antibiotics.
AMR mechanisms
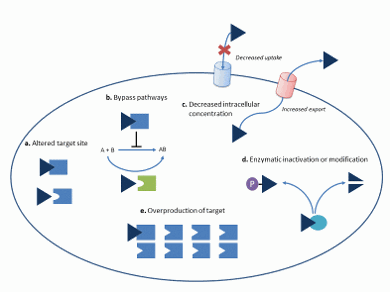
Antibiotics target essential parts of the bacterial metabolism, thereby killing them (bactericidal agents) or preventing them to grow (bacteriostatic agents). Typical mechanisms of action are inhibition of cell wall synthesis, inhibition of RNA or protein synthesis and inhibition of essential enzymes such as DNA gyrase, dihydropteroate synthetase or dihydrofolate reductase. As soon as bacteria are exposed to antibiotics, evolutionary pressure will promote the development of resistance. There are several ways in which resistance may develop (Fig. 2). From the diversity of resistance mechanisms, it is clear that evolution yields creative solutions, and that development of resistance should be taken into account in the development of novel antibiotics.
Development of novel antibiotics
The majority of the current antibiotics originates from microorganisms. Since 99% of the microbiome cannot (yet) be cultured, and many gene clusters have been discovered that code for unknown compounds, this represents a huge potential reservoir for novel antimicrobial compounds. The challenge is to further develop and apply technologies like synthetic biology, genomics and metabolomics for lead discovery. In the R&D and clinical trial pipeline, well over 90% of the candidate molecules fails and there is therefore an urgent need to fill the antibiotics pipeline with a high quantity and high quality of new drugs. Novel antibiotics should fulfill a number of criteria:
- high efficacy against the target microorganism, yet low liabilities for patients
- favourable chemical properties (stability, possibilities for derivatization)
- low resistance development (low resemblance to existing antibiotics)
To accomplish the aim of CARES, an interdisciplinary approach is needed, where biochemists, medical microbiologists and general microbiologists combine fundamental, basic science and translational science.