Deep imaging
A computer can look at, and learn from, many more images than a human specialist. AI systems are rapidly becoming indispensable for medical and biological applications. But they still have to learn how to explain their decisions.
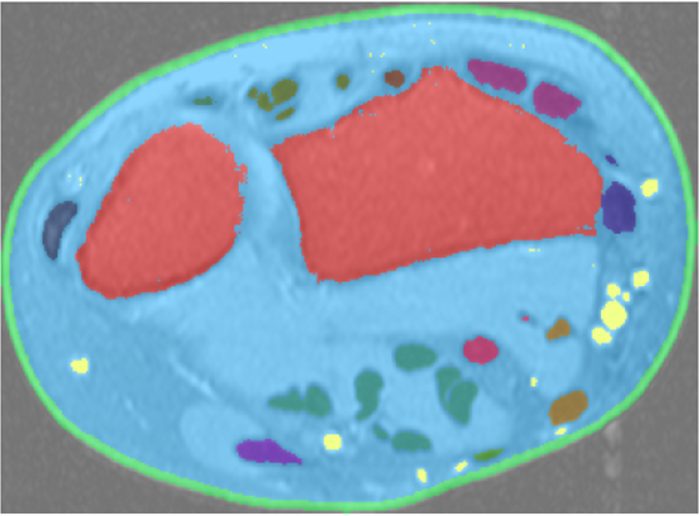
AI systems can identify tumours in a scan, but won't be making a diagnosis
X-ray images used to be clipped to a light box, and it was up to the radiologist to decide if a tumour, for instance, was visible on the image. Radiologists are pretty good at this. During training, they study thousands of such images while an experienced radiologist tells them whether that suspicious-looking spot is indeed a tumour.
An AI system, however, can look at a million X-rays to learn to identify tumours, and can, in theory, get better at this than any radiologist. The same goes for other imaging techniques, such as MRI and CT scans. According to Boudewijn Lelieveldt, Professor of Biomedical Imaging, the past five years have seen a revolution in this field: ‘Is automated image analysis new? No, but it has suddenly started to work much better.’ This is due to the availability of huge online photo databases, to fast graphics chips in computers and to deep learning networks.
Such networks can consist of dozens of layers. In the input layer, each cell registers one pixel of the image. The first inner layer detects an elementary feature, such as light and dark. The next layer might detect corners and edges in the image. Finally, the output layer can reliably draw contours around any tumours that are present.
Lelieveldt’s group recently investigated if training such a network, with data sets from different hospitals, made by scanners from different manufacturers, still delivers reliable results. This proved to be the case.
What does the patient stand to gain from this revolution? Until now, not every useful piece of information was extracted from a scan because this was too time consuming for medical specialists. Lelieveldt: ‘We have almost reached a stage where all these measurements can be done automatically.’ The comprehensive measurement and comparison of a set of scans is by no means a patient diagnosis. It is just one step in a process, and human doctors will remain in full control – for the present at least.
The concept of deep-learning networks will keep us busy for a further 20 years at least
MRI and CT scans are more or less at true scale, but Fons Verbeek, Professor of Computational Bio-Imaging, uses deep-learning networks to analyse microscope images. He collaborates a lot with biologists and with Naturalis Biodiversity Center, Leiden’s natural history museum: ‘They have one of the world’s largest collections of wood.’ He is working on an AI system that can identify the tree species from a microscope image of a piece of wood, and later even where this tree stood. This would make it much easier for customs officers to identify batches of illegal tropical hardwood.
Deep-learning networks are a success story, but they have weaknesses as well. When Verbeek tried to train a network to identify a certain type of individual cell under a microscope, it failed: ‘The network just threw everything into one basket. One cell against a nondescript background is just too boring for such neural networks.’
Nevertheless, a network with a more advanced architecture managed to do the job. The trick was to make some inner layers feed their output back into previous layers. This demonstrated once again that the deep-learning network is a very versatile concept and that it will keep computer scientists busy for a further 20 years at least. The possible variations in network architecture are almost infinite. Verbeek: ‘A few conventional architectures are considered standard at present. Sometimes you have to take a step backwards and play around a bit. There is still so much to explore.’

