Research project
Molecular basis of chromatin function
At the heart of the cell, in the nucleus, proteins and nucleic acids come together to maintain and express our genetic information. The interplay of proteins and nucleic acids in both genetic and epigenetic pathways forms the research focus in the Van Ingen group. How do their structures, motions and interactions come together to elicit function? The group’s main research line is centered on the molecular basis of chromatin function at the level of its repeating unit, the nucleosome.
Molecular basis of chromatin function
Chromatin is a highly dynamic system. Proteins continuously associate to and dissociate from chromatin, they place or erase epigenetic marks, they assemble and disassemble it when required, and they remodel its structure, all to ultimately control cell growth and differentiation. Malfunctioning of any of these processes underlies the onset of many diseases, including cancer. Unraveling the fundamental molecular principles of key steps in these processes is therefore required, not only for our understanding of chromatin biology in health, but also to provide a rational basis for future drug development.
To uncover the molecular mechanisms underlying chromatin function is a daunting task. The sheer size, and complexity and dynamics of the system form substantial challenges for any technology. One of the biggest challenges is the need to study the structural and dynamical changes in chromatin at atomic resolution, which is required for a mechanistic understanding. NMR is one of the few techniques that can bring this much-needed perspective. We thus use NMR as our main experimental technique, complemented by various biochemical, biophysical, and computational techniques, also in collaboration with other research groups.
Illuminating the nucleosome using methyl group based NMR
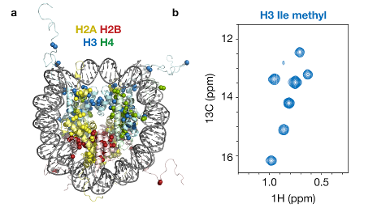 Fig 1. Methyl-based NMR of nucleosomes. (a) Crystal structure of the nucleosome (1KX5) highlighting observable methyl groups as spheres. (b) 13C, 1H-correlation spectrum for Ile-methyl groups in histone H3 inside the nucleosome.
Fig 1. Methyl-based NMR of nucleosomes. (a) Crystal structure of the nucleosome (1KX5) highlighting observable methyl groups as spheres. (b) 13C, 1H-correlation spectrum for Ile-methyl groups in histone H3 inside the nucleosome.The repeating unit of chromatin, the nucleosome, is a 200,000 Da supramolecular assembly of roughly one part DNA and one part protein. The massive size of the nucleosome calls for state-of-the-art NMR techniques that are tailored for such high-molecular weight systems. These techniques use the ultra-sensitive observation of methyl-groups in the proteins to produce beautiful high-quality spectra (Fig. 1, ref. 2, 3). This in turn allows us to use these methyl groups as probes to monitor the structure, dynamics and interactions of the nucleosome.
Structural basis of epigenetic recognition
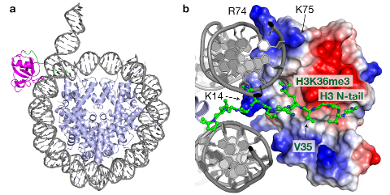 Fig 2. Structural basis of epigenetics: structural model, based on MNR and mutagenesis data, for the complex between the PWWP domain of reader protein PSIP1 and the nucleosome, trimethylated at H3K36. (a) Top view showing the PWWP domain in magenta and part of the H3 N-terminal tail in green. (b) Detailed view showing how the methylated lysine fits in the binding pocket of the PWWP domain, while at the same time an extensive basic surface (blue, key residues indicated)...
Fig 2. Structural basis of epigenetics: structural model, based on MNR and mutagenesis data, for the complex between the PWWP domain of reader protein PSIP1 and the nucleosome, trimethylated at H3K36. (a) Top view showing the PWWP domain in magenta and part of the H3 N-terminal tail in green. (b) Detailed view showing how the methylated lysine fits in the binding pocket of the PWWP domain, while at the same time an extensive basic surface (blue, key residues indicated)...The post-translational modification of histone proteins is a key mechanism in epigenetics. The majority of these chemical marks on chromatin serve as binding platforms for so-called reader proteins that in turn coordinate the functional state of chromatin. These protein-nucleosome interactions have in recent years become targets for the development of so-called ‘epigenetic drugs’.
The structural basis of reader-nucleosome interactions is an important focus of our research. Using tricks from chemical biology, we produce of nucleosomes that are decorated with specific epigenetic modifications. These tailor-made nucleosome can then be interrogated to reveal the molecular mechanism of epigenetic read-out.
Using this approach, we uncovered an unexpected role for the nucleosomal DNA in driving the recognition of an epigenetic mark (ref. 2). We determined the complex structure between a reader protein and a complete nucleosomes carrying an epigenetic mark at lysine 36 in histone H3 (Fig. 2). Importantly, this structure showed how the binding affinity of the reader protein for the trimethylated lysine is strongly enhanced by additional binding to the nucleosomal DNA, underscoring the potential of our studies to discover new concepts in chromatin biology.
Other interests
Integrative modeling of biomolecular assemblies. For many interesting systems, including nucleosome complexes, only sparse or low-resolution data may be available. Using integrative computational tools we can combine the data to generate meaningful atomic models of these systems.
Protein dynamics. Motions in proteins can be crucial for their function. Because of its sensitivity to motions on a very wide range of time scales, NMR is very well suited to not only study structure-function relationships, but also the role of dynamics herein.
NMR theory and methodology. As the saying goes, in theory there is no difference between theory and practice. In practice, this is only true for NMR. Well, at least practically true. NMR is essentially an applied form of quantum mechanics, allowing one to accurately design and simulate experiments.
Key publications
- Khan, F., M.A. Daniels, G.E. Folkers, R. Boelens, S.M. Saqlan Naqvi, H. van Ingen, "Structural basis of nucleic acid binding by Nicotiana tabacum glycine-rich RNA-binding protein: implications for its RNA chaperone function", Nucleic Acids Research, vol. 42, issue 13, pp. 8705 - 8718, 09/2014. DOI: 10.1093/nar/gku468
- van Nuland, R., F.M.A. van Schaik, M. Simonis, S. van Heesch, E. Cuppen, R. Boelens, H.T. Timmers, H. van Ingen, "Nucleosomal DNA binding drives the recognition of H3K36-methylated nucleosomes by the PSIP1-PWWP domain", Epigenetics & Chromatin, vol. 6, issue 1, pp. 12, 2013. DOI: 10.1186/1756-8935-6-12
- Kato, H., H. van Ingen, B.-R. Zhou, H. Feng, M. Bustin, L.E. Kay, Y. Bai, "Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR", Proceedings of the National Academy of Sciences, vol. 108, issue 30, pp. 12283 - 12288, 07/2011. DOI: 10.1073/pnas.1105848108
- van Ingen, H., F.M.A. van Schaik, H. Wienk, J. Ballering, H. Rehmann, A.C. Dechesne, J.A.W. Kruijzer, R.M.J. Liskamp, et al., "Structural Insight into the Recognition of the H3K4me3 Mark by the TFIID Subunit TAF3", Structure, vol. 16, issue 8, pp. 1245 - 1256, 8/2008. DOI: 10.1016/j.str.2008.04.015
- van Ingen, H., G.W. Vuister, S. Wijmenga, M. Tessari, "CEESY: Characterizing the conformation of unobservable protein states", Journal of the American Chemical Society, vol. 128, no. 12, pp. 3856-3857, Mar 29, 2006. DOI: 10.1021/Ja0568749
