Research project
Physicochemical analysis of allosteric binding pockets
Supervisor: Gerard van Westen
- Contact
- Gerard van Westen
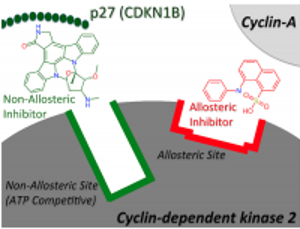
Allosteric modulators are ligands for proteins that exert their effect via a different binding site than the natural (orthosteric) ligand site and hence form a conceptually distinct class of ligands for a target of interest. Given that allosteric interactions tend to be non-competitive they are interesting from a drug design point of view as it enables the design of drugs that modulate rather than interfere with natural signalling pathways [1].
Recently we have established a data set of known allosteric interactions, which were gathered through text mining and the ChEMBL database) [2, 3]. This work was based on a cheminformatics approach and hence excluded the use of three-dimensional information in the form of crystal structures. However, previously Ritschel and coworkers have developed a fast pharmacophoric characterization tool of crystal structure binding pockets termed KRIPO [4].
This project is a collaboration with the authors of KRIPO and aims at characterizing both ligand properties of allosteric interactions and protein binding-site properties of allosteric interactions.
Requirements
Our research is interdisciplinary and we have both life scientists and computer scientists working in our group. Accordingly, most projects advertised in the context of the group are suitable for (MSc) students with either a chemical/biological/life science or a computer science/machine learning background. No previous experience in the other field is required, but interest to either get familiar with life science data, or with computational methods, would clearly be an advantage.We strongly support students to publish their results if possible and when the project results are suitable.
- Soudijn, W., I. van Wijngaarden, and A.P. IJzerman, Allosteric modulation of G protein-coupled receptors: perspectives and recent developments. Drug Discovery Today, 2004. 9: p. 752-758.
- van Westen, G.J., A. Gaulton, and J.P. Overington, Chemical, Target, and Bioactive Properties of Allosteric Modulation. PLoS computational biology, 2014. 10(4): p. e1003559.
- Gaulton, A., L.J. Bellis, A.P. Bento, et al., ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Research, 2012. 40(D1): p. D1100-D1107.
- Ritschel, T., T. Schirris, and F. Russel, KRIPO - a structure-based pharmacophores approach explains polypharmacological effects. Journal of Cheminformatics, 2014. 6(Suppl 1): p. O26.
